Cell culture-based production of influenza virus for vaccine manufacturing is faster and easier scalable than egg-based methods. In addition to being more stable, virus production in cell lines also gives higher yield and is safer to the end user. Influenza vaccines produced in mammalian cell lines require activation by trypsin. Trypsin is used to cleave the hemagglutinin (HA) protein on the influenza virus surface, making it infectious and enabling the virus to replicate in cell cultures. This process is crucial for efficient vaccine production in cell-based systems.
Our recombinant trypsin, TrypsiNNex® is a high-purity animal component-free cGMP trypsin that is easily integrated into existing and new protocols for influenza and other vaccine production. TrypsiNNex® has shown to efficiently propagate influenza virus production and increase virus infectivity.
Learn more from our application study on efficient cell dissociation using TrypsiNNex®:
An optimization study establishes a strategy for use of the new recombinant trypsin product TrypsiNNex® that boosts the propagation of H1N1 and H3N2 influenza viruses in MDCK-SIAT1 cells.
Summary:
This optimization study was conducted to establish an effective strategy for using TrypsiNNex® in cell culture-based influenza virus production. To that end, we devised a growth protocol for the MDCK-SIAT1 cell line and demonstrated its aptness as an in vitro system for propagating influenza viruses, especially with the exogenous trypsin TrypsiNNex®, which boosted viral particle production.
We employed RT-dPCR as a quick and efficient method to quantitatively assess virus yield during the optimization experiments. This method enumerates virus RNA copies with high sensitivity and correlates well with the more labor-intensive TCID50 assay. The latter, however, was instrumental in assessing the infectivity of produced virus particles.
The cornerstone of our investigation was a DoE screening and analysis to scrutinize the influence of 3 parameters – MOI, TrypsiNNex® concentration, and time until harvest – on the yield of virus particles, measured as RNA copies/mL. This meticulous analysis and a follow-up titration of TrypsiNNex® allowed us to pinpoint the following optimal production conditions:
- For H1N1 influenza virus: 005 MOI, 17 USP/mL TrypsiNNex®, harvest 3 days after infection
- For H3N2 influenza virus: 015 MOI, 1.5 USP/mL TrypsiNNex®, harvest 3 days after infection
Finally, applying these optimal infection conditions to production runs using TrypsiNNex® and 4 other commercial trypsin products, we demonstrated that TrypsiNNex® is a competitive reagent that boosts virus propagation on par with existing solutions. Moreover, based on TCID50 titers, TrypsiNNex® outperformed the alternative products in stimulating the propagation of biologically active, infectious influenza virus particles. TrypsiNNex® is a high-performance reagent that meets the standards of quality and non-animal origin raw materials for biopharmaceutical manufacturing.
The evolution of trypsin as a biopharmaceutical tool
The pancreatic serine protease trypsin occurs naturally in the digestive system of vertebrates. There, the inactive proenzyme trypsinogen is cleaved by enteropeptidase or trypsin itself to produce the active form.1 Well-characterized as a digestive enzyme since its first description in 1876 (2) trypsin hydrolyzes proteins, targeting primarily the carboxyl side of lysine and arginine in peptides (1).
The proteolytic activity of trypsin is an indispensable tool in both biological research and biopharmaceutical manufacturing. Trypsin is used in protein identification, quantification, and modification; in cell culture; and in tissue engineering. Demand for trypsin is significant and has predominantly relied on animal-derived trypsin. But more recently, safety regulations in biopharmaceutical manufacturing have made microbially expressed recombinant trypsin more viable than enzymes sourced from porcine or bovine pancreatic tissue.
TrypsiNNex®: a new animal-free recombinant porcine trypsin
Today’s stringent quality and safety measures for biopharmaceutical production encompass the careful selection of raw materials with minimal risk of harboring adventitious viruses. Despite rigorous testing, the infection of production cell cultures remains a significant concern with serious consequences. In the worst-case scenario, patients might be exposed to contaminated products. Less dire but also burdensome are production interruptions that cause drug shortages and extensive costs from investigations, corrective actions, manufacturing downtime, and regulatory scrutiny (3).
Raw materials of animal origin have an elevated risk of viral contamination. Data presented by Barone et al. 2020 show that materials testing alone fails to guarantee the absence of viruses in a product. Of 11 production contamination events examined, where raw materials were identified as the probable source, testing detected viral contamination of raw materials in only 3 cases (3).
TrypsiNNex® offers a solution to this issue. It aligns with industry standards for non-animal origin raw materials and reagents and essentially eliminates the potential for contamination by adventitious agents, proteases like chymotrypsin (5), and other impurities. Still, both animal-derived and most recombinant products exhibit autolytic activity that rapidly degrades the enzyme prior to purification and stabilization, leading to subpar performance (5).
TrypsiNNex® specifically addresses the need for a consistent, high-quality enzyme in biopharmaceutical manufacturing. This recombinant enzyme is expressed in Escherichia coli as an inactive protein and first becomes activated during the subsequent purification process. The short time interval between activation and stabilization curtails premature degradation and thus safeguards a substantial proportion of intact trypsin.
As a result, overall purity of TrypsiNNex® exceeds 90%, with over 70% β-trypsin and less than 20% α-trypsin (Figure 1). Deconvolution of the RP-HPLC spectrum in Figure 1 shows that the trypsin tails are derived from Na-adducts (data not shown), while other peaks are best characterized as product-related substances.
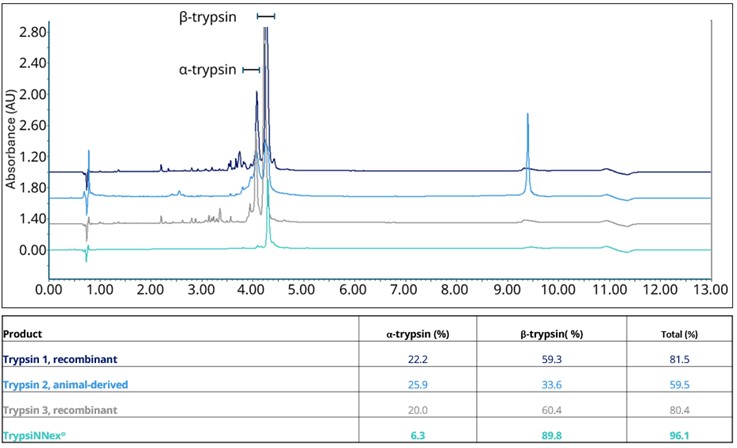
Trypsin in vaccine production: a balancing act
Trypsin is a key reagent in the production of viral vaccines, including influenza immunizations. Traditionally, influenza vaccines have been manufactured by inoculating embryonated chicken eggs, a labor-intensive and difficult-to-scale process that takes approximately 6 months to yield the required quantities (6). Therefore, in the last decades, the manufacturing of several influenza vaccines has transitioned to cell culture-based systems (7).
Cell culture-based methods use trypsin to activate influenza particles for the virus production. In the natural pathogenic process, the influenza virus binds to the host cell surface and triggers receptor-mediated endocytosis. The viral envelope then fuses with the endosomal membrane, releasing viral ribonucleoproteins into the cytoplasm. This fusion necessitates the proteolytic cleavage of hemagglutinin, which is induced by the low pH in the endosome and host proteases (8). For in vitro propagation of a virus, virus production, cultured cells do not harbor the necessary proteolytic activity, and thus, an exogenous protease is added to the cultivation medium. In vaccine production, trypsin is used to efficiently mediate activation and, thus, boost the propagation of viral particles (9) (10).
Finding the optimal amount of trypsin to promote virus propagation is a critical determinant of successful virus production in cell-based methods. Inadequate concentrations impede virus replication. Increasing trypsin concentration elevates virus output, but excessive amounts in the production medium can cause cell detachment (10). Achieving the right balance to devise a robust manufacturing process is facilitated by the use of a consistently high-performing enzyme like TrypsiNNex®.
Establishing optimal parameters for influenza virus propagation with TrypsiNNex®
Designed to circumvent the degradation issues observed for other trypsin products, TrypsiNNex® has a higher proportion of intact enzyme and a unique performance profile. Thus, we undertook optimizing its use in coordination with other parameters to maximize the output of influenza virus production without hampering cell function.
This application note summarizes the results of our optimization study and demonstrates the suitability and optimal use of TrypsiNNex® for influenza virus propagation in Mardin-Darby Canine Kidney (MDCK) cell cultures. We used a Design of Experiments (DoE) approach to scrutinize the influence of multiplicity of infection (MOI), TrypsiNNex® concentration, and harvest day on the yield of H1N1 and H3N2 influenza virus strains. With a validated cell culture protocol and quantified viral stocks, we determined optimal virus production conditions and conducted a comparative analysis to benchmark the performance of TrypsiNNex® against other products on the market.
A validated cultivation protocol for MDCK-SIAT1 cells
MDCK cells are widely recognized as a standard producer cell line. They express sialic acid surface receptors that serve as entry points for influenza viruses. The engineered MDCK-SIAT1 cell line chosen for this study includes a human CMP-N-acetylneuraminic: β-galactoside α-2,6-sialyltransferase (SIAT1) transgene that drives an overexpression of these receptors. The cells exhibit enhanced influenza propagation rates and higher titers than the parental cell line (11) (12).
To establish growth conditions that rapidly generate healthy, confluent cell cultures for virus inoculation, we examined the growth profiles of MDCK-SIAT1 cells. This analysis aimed at determining population doubling rates, optimal seeding concentrations, and appropriate passaging intervals. Frozen cells from passage 8 were thawed, suspended in a growth medium consisting of DMEM, 10% FBS, 0.2% BSA, 25 mM HEPES, 100 USP/mL penicillin/streptomycin solution, and 1 mg/mL of G418, and then incubated in T75 flasks at 37°C and 5% CO2. The G418 antibiotic served to select for the engineered cells, as they carried a corresponding resistance gene.
The cells grew slowly after thawing (29 h) but maintained shorter and relatively constant doubling times in subsequent passages (14.8 h, on average). We tested different seeding cell densities between 5×103 and 1.47×104 cells/cm2, which influenced how quickly the cells reached 90–100% confluency (Figure 2). Generally, the cultures had a cell density of 1.4–1.7×105 cells/cm2 after 2–3 days of cultivation. Subsequent analyses (not shown) confirmed that cells were healthy and displayed an optimal physiological state at this density.
To ensure that growth was not limited by nutrient depletion or metabolite accrual, we examined spent media from passage 10. The analysis revealed low L-glutamine levels and high amounts of lactate, which could restrict cell reproduction. Therefore, we raised the volume of medium added to each T75 flask in subsequent passages from 20 to 30 mL. At passage 12, the cells were also allowed to grow until day 4. While doubling times did not change, the spent media harvested on day 4 had limited L-glutamine and excess lactate. Thus, a fourth day of cultivation provided no benefit.
Based on these results, we established the following as optimal cultivation conditions for MDCK-SIAT1 cells:
- A seeding density of approximately 5×104 cells/cm2 with a 2-day cultivation period was selected for all experiments.
- Incubation at 37°C with 5% CO2 in 30 mL of growth medium for a T75 flask supported unrestricted cell growth and healthy cells.
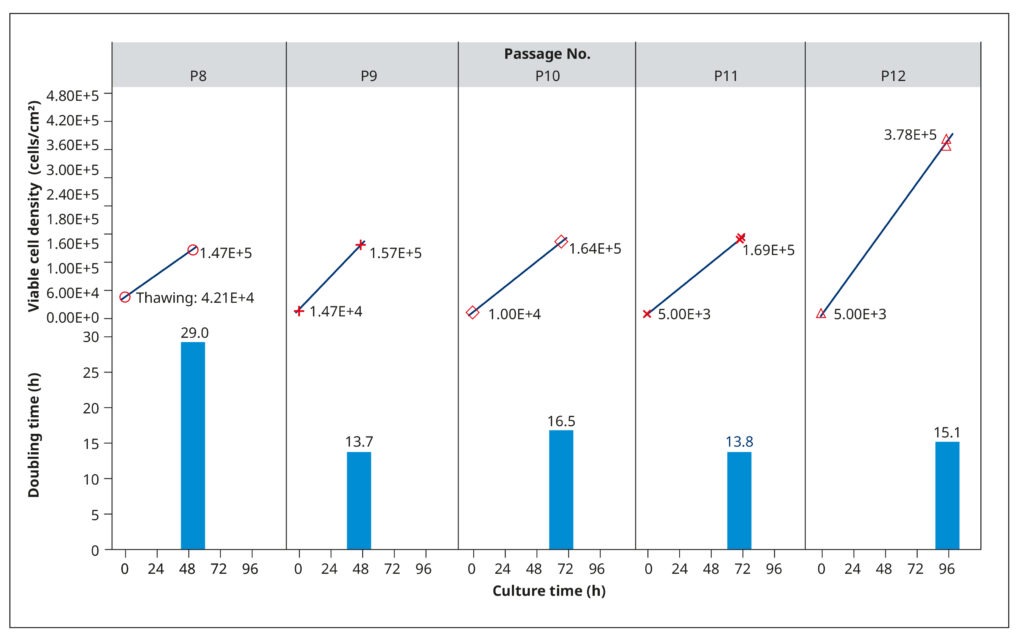
Production and quantification of suitable working virus stocks
We established suitable working stocks of H1N1 and H3N2 influenza virus to carry out all optimization experiments. MDCK-SIAT1 cells were seeded and cultured in T75 flasks. On the day of infection, the cells were near optimal confluence at a density of 1.29×105 cells/cm2 and exhibited normal morphology. We inoculated each T75 flask with 600 µL of the original virus seeds and then incubated the cultures for 30 minutes at 37°C and 5% CO2 to facilitate virus adsorption.
Following the adsorption period, the cells were washed with Dulbecco‘s phosphate-buffered saline and covered with 20 mL of serum-free virus growth medium (DMEM, 2 mM L-glutamine, 0.2% BSA, 25 mM HEPES, and 100 USP/mL penicillin/streptomycin solution) containing 7.0 USP/mL of TrypsiNNex®. This specific TrypsiNNex® concentration was found to generate viral titers comparable to those obtained with trypsin treated with L-1-tosylamido-2-phenylethyl chloromethyl ketone (TPCK-treated; data not shown). After 3 days of virus propagation at 37°C and 5% CO2, the supernatant was collected, centrifuged, and examined. The majority of inoculated cells were no longer viable, suggesting that longer cultivation would not significantly increase virus yield. One-milliliter aliquots of virus-containing supernatant frozen at –80°C were our working virus stocks.
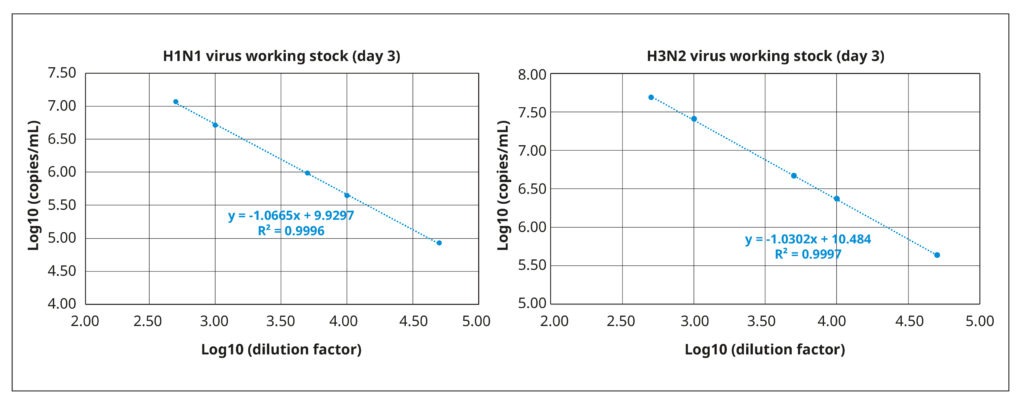
We quantified viral titers of both stocks via reverse transcription coupled with digital PCR (RT-dPCR). RT- dPCR is a highly sensitive and reproducible method for quantifying copies of viral RNA molecules that, in our experience, is superior to quantitative PCR. Briefly, RNA was extracted from 140 µL working virus stock using the QIAamp® Viral Mini Kit (QIAGEN®), and the RT-dPCR was performed with the QIAcuity® OneStep Advanced Probe Kit (QIAGEN®) in combination with gene- specific Taqman probes and primers. Fast and with high throughput, RT-dPCR was ideal for the repeated quantifications of our optimization experiments. It also has a broad dynamic range (2 logs), as demonstrated by the strong correlation between amplification results and serially diluted samples of the working stocks (Figure 3).
The working stock of the H3N2 virus had a titer of 2.37×1010 RNA copies/mL, whereas the working stock of H1N1 virus had a lower virus particle concentration of 4.92×109 RNA copies/mL.
Low multiplicity of infection and harvest after 3 days are predicted to maximize viral titer
With a validated cell culture system and viable viral stocks, we employed a DoE approach to investigate the influence of MOI, TrypsiNNex® concentration, and harvest day on the propagation of each influenza strain.
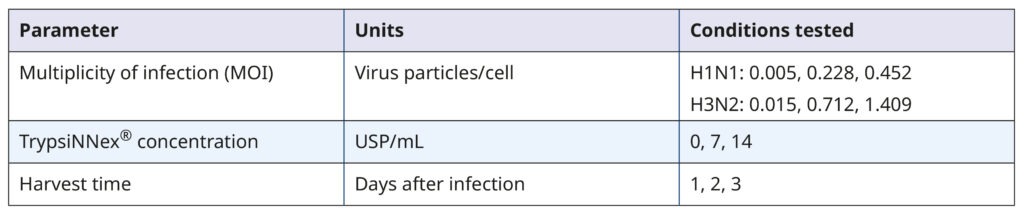
Our goal was to identify conditions that maximize viral titer. Table 1 summarizes the experimental parameters and their tested values.
Table 1. Parameters and values tested in the DoE screening
The DoE experiments combining all permutations of the 3 parameters were done on 6-well plates. MDCK- SIAT1 cells were first seeded and allowed to grow for 2 days. On the day of infection, each well contained approximately 1.93×106 cells. Working viral stocks were added to the cells to achieve the target MOI based on their infectious titer (in TCID50/mL, not shown) and allowed to adsorb to the cell monolayer for 30 minutes. When added stock volumes were less than 200 µL, we supplemented them with viral growth medium to ensure uniform coverage in all wells. After washing the cells, we added 2 mL of viral growth medium with the target TrypsiNNex® concentration to each well and incubated the plates for virus propagation. Supernatants were collected, centrifuged, aliquoted, and frozen on the specified harvest days. The viral titer of each harvest was quantified by RT-dPCR, and the values were fed into the DoE analysis using JMP.
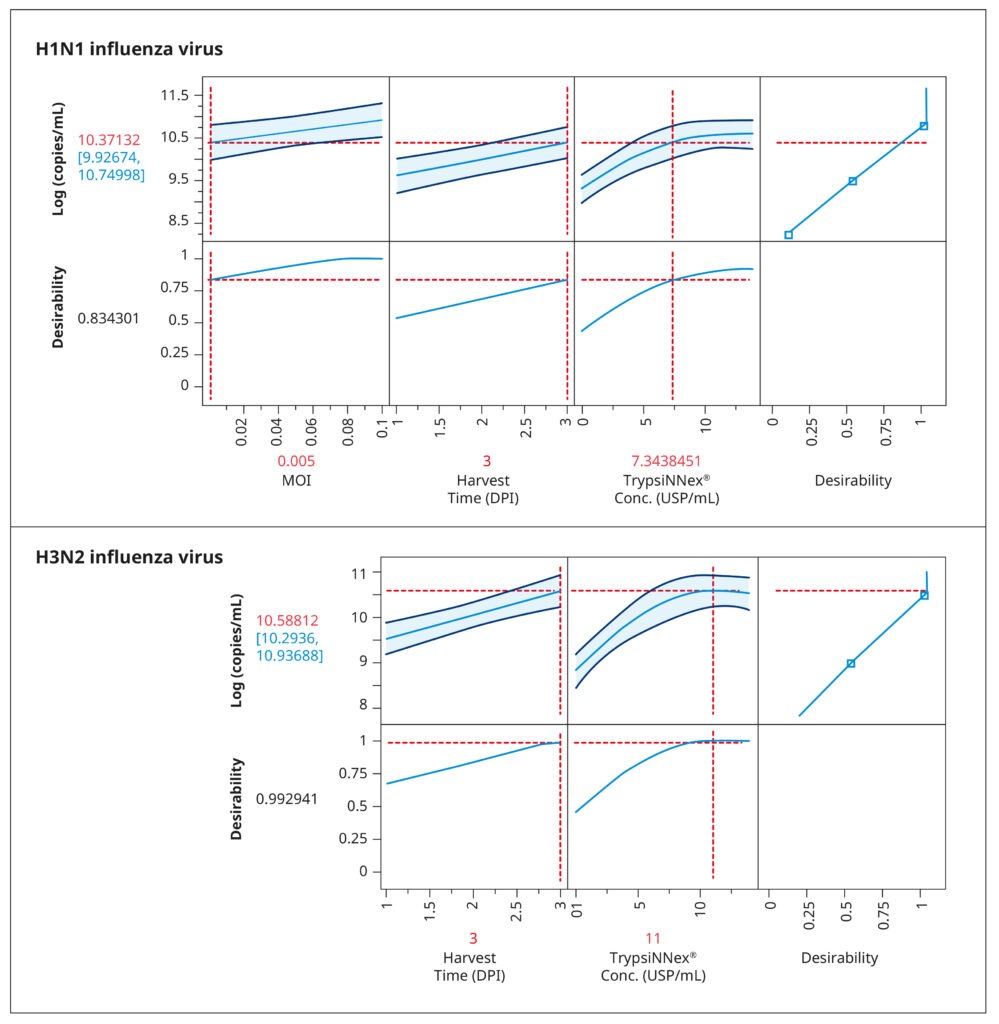
The resulting model elucidated the influence of the tested parameters and their interactions on viral titer. Figure 4 visually represents the model‘s predictions. The parameter values depicted in red were predicted to maximize viral titer. While an MOI of 0.005 viral particles per cell enhanced H1N1 influenza production, MOI had no substantial impact on H3N2 influenza. For both strains, the optimal harvest time was 3 days post- infection, with H3N2 influenza benefiting from a higher concentration of TrypsiNNex® (11 USP/mL) compared to H1N1.
To confirm the predicted optimal conditions, we conducted a follow-up experiment comparing them to conditions that yielded the next best viral titer levels in the DoE (Table 1). The comparison reaffirmed the optimal conditions for H1N1 influenza production as a low MOI of 0.005, 7 USP/mL TrypsiNNex®, and a 3-day harvest time. For H3N2 influenza, our results corroborated that a high MOI did not improve infection efficiency or final viral titer. Thus, using a low MOI of 0.015 is an efficient and resource-saving strategy.
Also the 3-day harvest time was shown to produce the highest H3N2 titers. The only discordant outcome pertained to TrypsiNNex® concentration, where 14 USP/mL yielded a slightly better viral titer than the predicted 11 USP/mL. Therefore, we proceeded to fine- tune TrypsiNNex® concentration using the identified strain-specific optimal values for MOI and harvest day.
Different TrypsiNNex® concentrations maximize the production of H1N1 and H3N2 influenza strains
To evaluate more systematically the influence of TrypsiNNex® concentration on the yield of H1N1 and H3N2 viral particles, we conducted a series of production runs using TrypsiNNex® concentrations from 0 to 17 USP/mL. We maintained the optimal MOI and harvest day identified in the DoE analysis across all production runs and adhered to the same protocol as in the infection experiments of the DoE screening.
This time, however, we quantified the viral titers of the harvested supernatants by 2 methods: RT-dPCR and a tissue culture infectious dose assay (TCID50). TCID50 is a widely used virus titration assay that gauges titers based on the cytopathic effect of a virus-containing solution on a host cell culture. Specifically, it reports the dilution of a virus required to infect 50% of the cell culture. While TCID50 is commonly used as a measure of the biological activity of a virus, it has practical drawbacks: it is a non-quantitative measure that relies on cell staining scores and involves a labor- intensive, multi-step process that spans 2 weeks. Our in-house comparisons have demonstrated that RNA copy enumeration by RT-dPCR and infectious titer measured as TCID50/mL correlate well. Consequently, the reproducibility and efficiency of RT-dPCR made it an ideal method to quantify physical titers for our process optimization, while TCID50 was valuable for confirming virus function.
Figure 5 illustrates the results of our TrypsiNNex® concentration experiments. Overall, adding TrypsiNNex® to the virus-inoculated MDCK-SIAT1 cells boosted the propagation of both H1N1 and H3N2 viral particles.
Compared to no TrypsiNNex®, virus titers measured by RT-dPCR increased 10 and 100-fold, respectively, with the addition of 1.5 USP/mL TrypsiNNex®. However, the impact of more TrypsiNNex® diverged between the H1N1 and H3N2 influenza strains. With H1N1, elevating the TrypsiNNex® concentration beyond 1.5 USP/mL positively correlated with higher viral titer levels. In fact, 17 USP/mL TrypsiNNex® produced the highest physical titer accompanied by a peak in functional titer (Figure 5, left). In contrast, concentrations of TrypsiNNex® above 1.5 USP/mL suppressed viral titers of H3N2 in the harvested supernatant. 1.5 USP/mL TrypsiNNex® yielded the highest physical and functional titer for H3N2 virus particles (Figure 5, right).
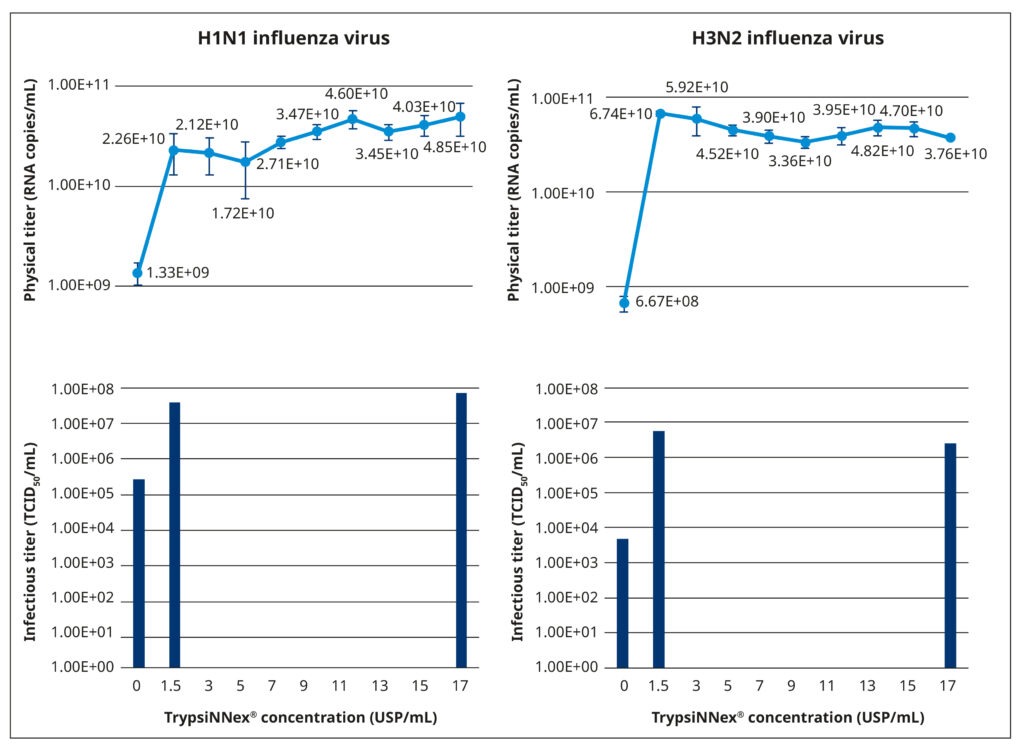
Based on these results, we adopted the identified strain-specific TrypsiNNex® concentrations for the following set of performance equivalency experiments; that is, 17 USP/mL TrypsiNNex® for the production of H1N1 influenza virus and 1.5 USP/mL for the production of H3N2 influenza virus.
TrypsiNNex® is a high-quality alternative to other trypsin products
To benchmark the performance of TrypsiNNex® in virus production and ascertain its suitability for clinical applications or as an alternative research reagent, we compared it with 4 existing trypsin products in a performance equivalency study. These products included a TPCK-treated bovine trypsin, trypsin derived from porcine pancreatic tissue, and 2 recombinant enzymes.
We evaluated the 5 trypsin products under identical conditions, using the same cell culture system, working viral stocks, and the strain-specific MOI, harvest time, and trypsin concentrations defined in our optimization study. The trypsin concentration was normalized to the activity of each product. As in the protocol of the DoE screening, MDCK-SIAT1 cells were permitted to reach the desired monolayer density of 1.4–1.7×105 cells/cm2 and then inoculated with 200 µL working virus stock at the appropriate MOI. After a 30-minute incubation at 37°C and 5% CO2 to facilitate virus adsorption, the cells were carefully washed and covered with 2 mL of virus growth medium containing the test trypsin. The cells were then incubated at 37°C and 5% CO2 for 3 days, and the supernatant was harvested, centrifuged, and stored at –80°C. Physical (RNA copies/mL) and functional (TCID50/mL) titers were measured for all harvests.
All tested trypsin products yielded physical titers within the same order of magnitude (Figure 6). One notable exception was trypsin extracted from porcine pancreatic tissue, which produced a significantly lower physical titer for the H1N1 strain. Among the tested products, TrypsiNNex® did not produce the highest virus yield based on physical titer measurements, falling approximately 25–50% below the peak yield for each strain. However, under these specific production conditions, TrypsiNNex® delivered the highest functional titers for both influenza strains. The H1N1 and H3N2 virus particles produced using TrypsiNNex® exhibited enhanced infectivity (at least 13% and 84% higher, respectively) compared to viral particles propagated with the other trypsin products.
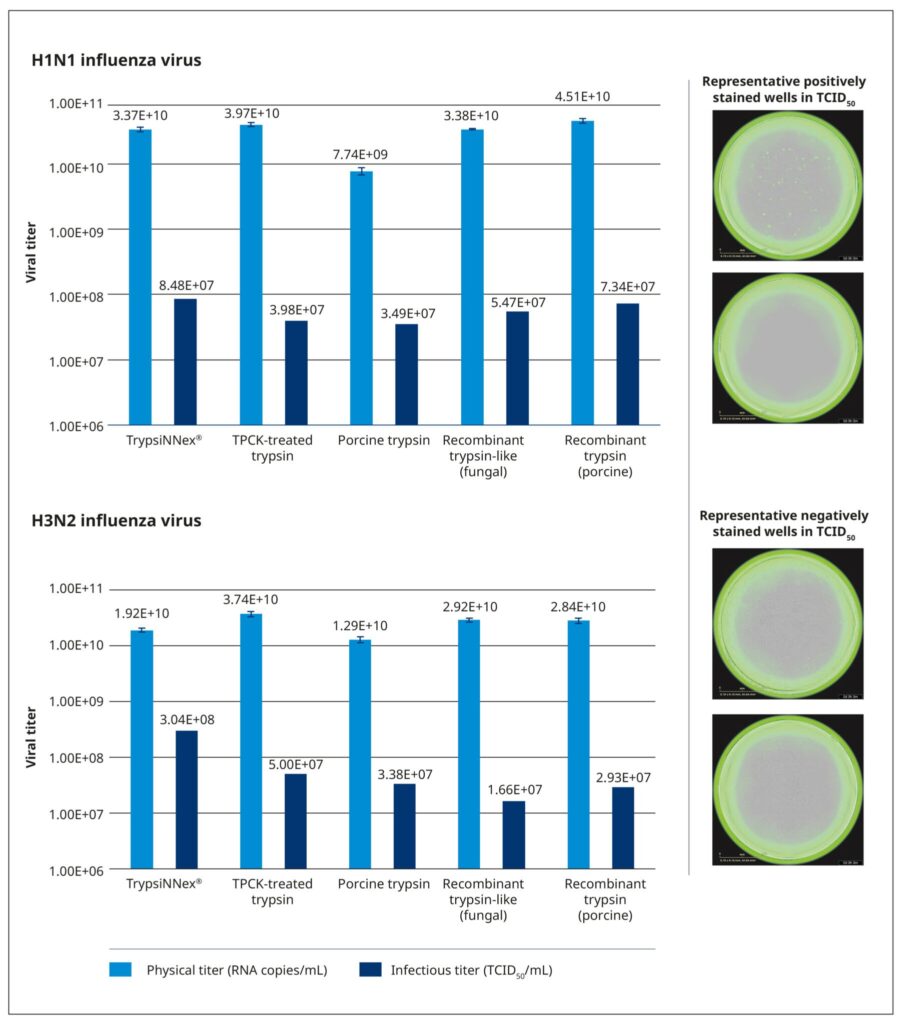
References
Rawlings, D. and Barrett, A.J. 1994. Families of serine peptidases. Methods in Enzymology 244: 19. doi:10.1016/0076-6879(94)44004-2
Kühne, 1876. Ueber das Trypsin (Enzym des Pankreas). Verhandlungen des Naturhistorisch- Medicinischen Vereins zu Heidelberg 1: 194.
- Barone, W. et al. 2020. Viral contamination in biologic manufacture and implications for emerging therapies. Nature Biotechnology 38: 563. doi: 10.1038/s41587-020-0507-2
- European Medicines 2014. Guideline on the use of porcine trypsin used in the manufacture of human biological medicinal products. https://www.ema.europa.eu/en/use-porcine-trypsin-used- manufacture-human-biological-medicinal-products-scientific-guideline (accessed September 2023).
- Heissel, et al. 2019. Enhanced trypsin on a budget: stabilization, purification, and high-temperature application of inexpensive commercial trypsin for proteomics applications. PLoS ONE 14: e0218374. doi: 10.1371/journal.pone.0218374
- Centers for Disease Control and Prevention. Selecting viruses for the seasonal influenza vaccine. https://www.cdc.gov/flu/prevent/vaccine-selection.htm(accessed September 2023).
- Audsley, M. and Tannock, G.A. 2008. Cell-based influenza vaccines. Drugs 68: 1483. Doi: 10.2165/00003495-200868110-00002
- Böttcher-Friebertshäuser, et al. 2013. Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium. Pathogens and Disease 69:87. doi: 10.1111/2049- 632X.12053
- Klenk, D. et al. 1975. Activation of influenza A viruses by trypsin treatment. Virology 68: 426. Doi: 10.1016/0042-6822(75)90284-6
- Genzel, Y. et al. MDCK and Vero cells for influenza virus vaccine production: a one-to-one comparison up to lab-scale bioreactor cultivation. Applied Microbiology and Biotechnology 88: 461. doi: 10.1007/s00253-010-2742-9
- Oh, D.Y. et al. MDCK-SIAT1 cells show improved isolation rates for recent human influenza viruses compared to conventional MDCK cells. Journal of Clinical Microbiology 46: 2189. doi: 10.1128/ JCM.00398-08
- Matrosovich, et al. 2003. Overexpression of the alpha-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. Journal of Virology 77: 8418. Doi: 10.1128/ jvi.77.15.8418-8425.2003
- Lei, et al. 2021. On the calculation of TCID50 for quantitation of virus infectivity. Virologica Sinica 36:doi: 10.1007/s12250-020-00230-5
- Lugovtsev, Y. et al. 2013. Heterogeneity of the MDCK cell line and its applicability for influenza virus research. PLoS ONE 8: e75014. doi: 10.1371/journal.pone.0075014
Download application note
Request quotation
Request a sample